-
The CGH module is a system capable of detecting quantitative (unbalanced) genome changes (amplification / deletion).
-
LUCIA Cytogenetics can work with fluorescent probe kits from different manufacturers and with home-made kits as well.
-
Get high-quality images using scientific digital CCD cameras with 14 bits analog-to-digital conversion.
-
Perfect linearity of the camera ensures that the results of the analysis are not deformed.
-
Maximum possible image information is transferred to the computer with no loss of details thanks to the automatic camera exposure control.
-
The completely software-controlled motorization of the microscope speeds up image acquisition and reduces the risk of human-caused errors.
-
Any combination of the individual color images can be displayed as one composed image.
-
Create karyograms using the full-featured karyotyping module.
-
Check and correct chromosome axes manually to get even better results.
-
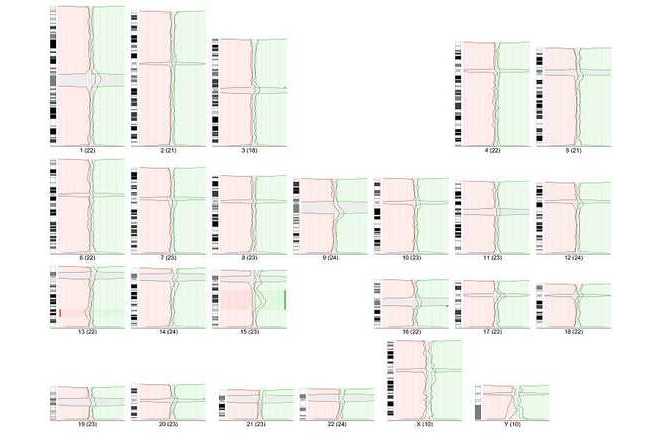
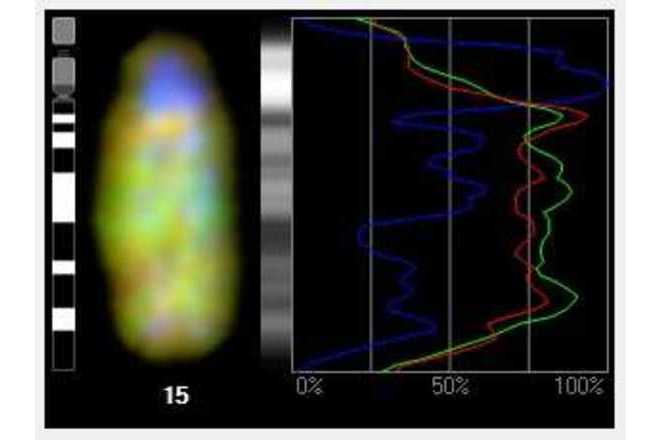
Use profile graphs to study intensity profile of each individual color channel.
With the CGH module you can:
-
Use standard CGH or high resolution CGH.
-
Analyze multiple images of the same sample to get reliable results.
-
Manage the reference sets for high resolution CGH comfortably.
-
Check the influence of each individual chromosome and exclude it from the result if needed.
-
Set various user-defined analysis and display parameters.
-
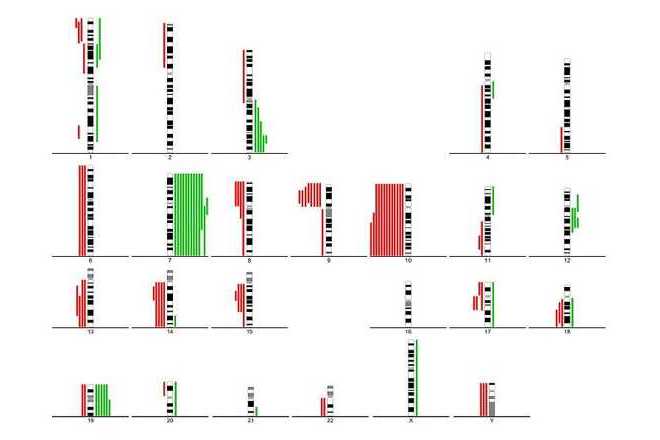
Use summary lines to indicate DNA gains and losses.
The CGH module is fully compatible and can be integrated with other LUCIA Cytogenetics modules:
-
Report Tool including all CGH display options and CGH summary graphs.
Comparative genomic hybridization (CGH) is a molecular-cytogenetic method based on fluorescence in situ hybridization (FISH) which is able to detect quantitative (unbalanced) genome changes (amplification / deletion). This method is very useful for examination of tissues which cannot be cultivated easily or in cases where viable cells are not available (solid tumours, hematological malignancies, etc.). This method is based on the in situ hybridization of total genomic DNA from subject tissue (tested material) and DNA from normal control tissue (reference), each labeled differently, and subsequent hybridization to normal metaphase chromosomes. Copy number variation is detected by measuring regional differences of the tested vs. reference fluorescence intensity ratio for each chromosome locus.
This technique only detects imbalances (gain or loss of DNA) in the genome. Balanced rearrangements such as translocations, inversions and other aberrations, which do not change the copy number, can not be detected.
Standard chromosomal CGH can detect changes that are present in 30 – 50 % or more of the specimen cells and where size of the chromosomal region affected exceeds 5-10 Mb. For improving specificity and sensitivity of the method, high resolution CGH using advanced statistics can be applied.